Testing
As the season draws to a close there is time to take stock of the season past and make plans for the coming year.
There are many things that can be done to ensure that the crops we grow are as healthy (competitive) as possible. These include;
- Testing retained seed for seed borne diseases.
- Collecting seed samples for herbicide resistance testing.
- Sampling for soil borne diseases.
Seed testing and dressings key to smut management
Western Australian barley growers concerned about levels of loose smut in their crops this year are encouraged to have their saved barley seed tested for the disease after harvest and to apply seed dressings next season.
Barley growers, particularly in medium to high rainfall areas, experienced high levels of barley loose smut (Ustilago nuda) this year. A long, cool spring in 2016 led to high infection rates of seed used to sow 2017 barley crops. This year’s spring has also been relatively favourable in many barley growing areas, so smut levels might be high again next year.
Department of Primary Industries and Regional Development (DPIRD) researcher Andrea Hills has been researching loose smut as part of the Western Pathology project (Improving grower surveillance, management, epidemiology knowledge and tools to manage crop disease) co-invested by Grains Research and Development Corporation (GRDC).
Growers of Hindmarsh and La Trobe, are more likely to be affected by the disease, which reduces grain yield and affects export opportunities for some markets.
Testing seed retained at harvest would give growers a better idea of what they might face if they did not treat it carefully with a premium seed dressing. The results of product efficacy trials are available at Controlling barley loose smut.
If seed is tested and found to be near the 5 per cent infection level, it may be better to replace the seed lot with a new batch of seed, – for example sourced from the lower rainfall area which usually has lower levels of smut. Certified seed has a limit of 0.05% loose smut.
To keep smut under control, growers should use seed dressings every year and a thorough application is critical, with every seed needing a dose; when treating seed, slow the auger down and keep water rates high.
It is not possible to prevent seed from becoming infected with loose smut, and seed dressings can only control the smut that is already present in the grain and while no seed dressing will give 100 % control treating seed is still worthwhile and stops the problem from escalating.
In 2013 DPIRD trials showed that some premium seed dressings controlled the disease at the 99 per cent level. At a paddock scale, even with 99.9 per cent control and perfect application, at a 70kg/ha sowing rate of ‘average’ La Trobe sized seed that has a 1 per cent level of infection, there will still be 1.4 infected plants for every 30 by 30 metre square.
More information about barley loose smut management is available by searching the DPIRD website.
Growers wishing to test seed can contact DPIRD Diagnostic Laboratory Services (previously AGWEST Plant Laboratories) on 08 9368 3351 or ddls@agric.wa.gov.au.
Field pea - Pea seed-borne mosaic virus guide for preparing for 2018
In preparation for the 2018 growing season, pea growers should have their seed tested for Pea seed-borne mosaic virus (PSbMV) infection, especially if harvested seed from previous years is to be sown. A cumulative effect of seed infection often occurs with PSbMV. If buying in seed from a seed company, growers should check that this test has been done and if not have the seed tested themselves. The DPIRD Diagnostic Laboratory Services (DDLS) can perform the test with a 500g sample of the pea seed (details below).
PSbMV is a seed-borne pathogen that is spread between pea plants by winged migrants of a multitude of aphid vector species originating outside of the crop. The virus causes canopy depressions consisting of stunted plants. The disease caused by PSbMV infection can cause significant seed yield losses and have detrimental effects on seed quality including seed deformation and dark ‘tennis-ball’ markings. Brenda Coutts from DPIRD conducted research in 2005 and 2006 examining the effects of sowing field pea seed with different amounts of PSbMV infection on virus spread, seed yield, seed quality and infection levels in harvested seed. Plots were sown with actual or simulated seed transmission rates of 0.3%-6.5% in 2005 and 0.1%-8% in 2006 and spread was by naturally occurring migrant aphids. In these trials, PSbMV-induced seed yield and quality losses increased with the amount of primary inoculum present, reaching >25% when high levels of spread occurred prior to flowering.
Furthermore, PSbMV seed transmission rates to harvested seed increased as incidence increased, but decreased in situations where spread was minimal. From this information and an epidemiological study produced in 2016, a combination of pre-sowing rainfall and seedborne inoculum were identified as primary drivers of PSbMV spread and subsequent seed yield and quality losses.

A forecasting model for PSbMV has been developed as part of an ARC linkage PhD project and co-funded by the DPIRD led GRDC National plant pathogen modelling project. The model informs a decision support system that provides location specific PSbMV risk forecasts prior to sowing This allows sufficient time to implement control recommendations, such as having seed samples tested for PSbMV transmission rate to seedlings, obtaining seed with minimal infection or of a PSbMV resistant cultivar, and implementation of cultural management practices.
Each season, growers can be reminded to get seed tested, assess their PSbMV risk and make informed management decisions using the free SMS PSbMV risk alert service. This risk alert service will complement the existing blackspot risk alert.
If you would like to subscribe to this service please text ‘pea virus’ to +61 (0)475 959 932 with your name and closest weather station or your locality.
Management
As well as sowing healthy seed, an integrated management approach is vital to reducing losses since insecticides are ineffective for PSbMV control;
- Target early canopy development. Early seeding at high rates (more than 120kg/ha) and narrow row spacing (18cm) will promote an early, dense plant canopy before aphid arrival. This will help shade out the seed-infected plants that act as a source of PSbMV infection and also reduce aphid landings. In 2007, research by Brenda Coutts (DPIRD) found that sowing higher seeding rates with straw mulch and regular insecticide application resulted in slower spread and lower seed infection than sowing at a standard seeding rate without straw mulch and or insecticide.
- Retaining stubble helps decrease early virus spread by reducing aphid landing rates while bare earth between plants attracts incoming migrant aphids.
- Avoid potential virus sources by isolating pea crops from other pulse or field pea crops sown with untested seed stocks as these could be a potential source of PSbMV.
- Cultivar choice. If healthy seed of the agronomically best cultivar is not available, use PSbMV-resistant Wharton.
Herbicide resistance testing
Herbicides play a major, and expensive, role in our farming systems. The continued usefulness of many of our common herbicide groups is under serious threat from the development of herbicide resistance in weed populations. Testing weeds for their susceptibility to herbicides enables informed decisions to be made in relation to herbicide application and choice, and potentially save money by avoiding herbicides that are no longer effective.

Unfortunately, only a small percentage of growers test herbicide efficacy and this is often in response to a herbicide failure. There are two major tests; Quick-Tests™ for testing seedlings and small plants during the season and seed testing at the end of the season.
Harvest time is the perfect time to collect seed of weeds suspected of having resistance to herbicides.
Once the seeds are collected from the paddock, they are sent to the lab where they are grown and sprayed with the specified herbicides to give answers relevant to your farming system. Results are normally available 3-4 months after harvest.
Further information on the best way to collect and package collected seed samples is available on the herbicide resistance and susceptibilty testing webpage along with links to the testing labs.
The Australia Herbicide Resistance Initiative (AHRI) conducts regular surveys of the grainbelt that investigate the level and extent of resistance to a range of herbicide groups for a number of weed species. The surveys are random and conducted roughly every 5 years.
The AHRI website provides the survey results in an easily searchable format so that specific weed/crop and region interactions can be explored and made more personal to the grower. Click here to go to the survey results.
Testing for glyphosate resistance
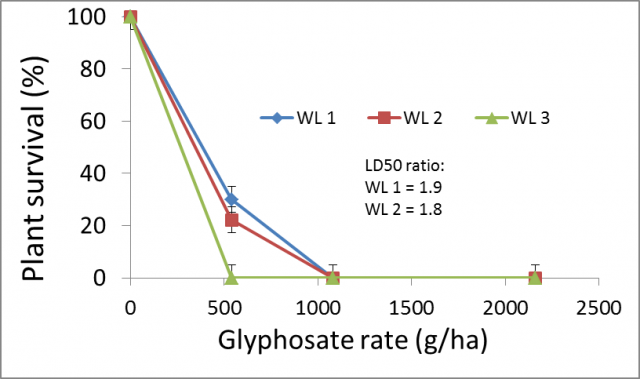
Dr Abul Hashem (DPIRD) has been conducting regular surveys of roadside and summer weeds as part of a GRDC supported project. Various plant populations have also been subjected to herbicide resistance testing as part of this process. Recently a population of willow leaf lettuce (Lactuca saligna) has been found in the Gascoyne region that is resistance to glyphosate. While this weed species is not necessarily of importance to growers in the grainbelt, it is closely related to weeds such as sowthistle and prickly lettuce that do occur in the grain growing regions.
Three willow-leaved lettuce populations were tested with three rates of glyphosate, applied at the 3-4 leaf stage (Figure 1). 1000g/ha glyphosate was required to kill all three populations.
Characterising soil borne disease risk in the eastern wheatbelt
During the 2017 season, Kylie Chambers (DPIRD Merredin) conducted a survey to investigate the presence and incidence of disease in the eastern wheatbelt. The survey was funded by the National Nematology project with investment from GRDC.
During March- June 2017, PreDicta B® samples were taken to provide a snapshot of the disease inoculum in cropping paddocks around the eastern wheatbelt (Kwinana East). A total of 136 samples from 128 paddocks were analysed using the PreDicta B® test from sites across the eastern wheatbelt.
Disease inoculum levels were sorted into high, medium, low and not detected categories based on the SARDI risk and population calibrations.
The percentage of each category was determined for the tests in each shire to indicate the overall risk/ population in the eastern wheatbelt by shire.
Several diseases were not detected in the survey including CCN, ascochyta blight, phytophthora root rot, black spot (Phoma Koolunga) and the nematode Pratylenchus thornei.
Three major pathogens known to cause significant crop loss were frequently detected in the survey: crown rot, rhizoctonia and the root lesion nematode Pratylenchus neglectus (Table 1). The nematode Pratyenchus penetrans was only detected in one sample in the Tammin shire and P. quasitereoides detected twice at low levels in the Merredin shire.
| Pathogen | High disease risk | Medium disease risk | Low disease risk | Not detected |
| Crown rot | 36% | 8.1% | 17.7% | 38.2% |
| P. neglectus | 5.1% | 51.5% | 22.1% | 21.3% |
| Rhizoctonia | 3.7% | 16.2% | 29.4% | 50.7% |
* risk categories are a guide only, may be subject to regional and seasonal differences, and may be revised over time.
Symptoms of crown rot, rhizoctonia and root lesion nematode have been detected in the eastern wheatbelt this season.
Take-all was also detected in 50.7% of samples (21.0% medium; 29.7% low disease risk levels), although is thought to be of lesser consequence in the eastern wheatbelt due to the lower levels of rainfall.
Other diseases such as common root rot (Bipolaris) and pythium root rot were also found at a high frequency, however, disease risk calibrations have not yet been developed for these diseases (Table 2).
| Pathogen | Detected | Not detected |
| Common root rot | 53.4% | 46.6% |
| Pythium root rot | 72.1% | 27.9% |
Root diseases impacts can be difficult to recognise and targeted testing of paddocks can identify situations of increasing or high risk. While PreDicta B® quantifies the level of pathogen DNA in the soil as an indicator of disease risk, it does not predict the disease severity that will occur as this will also be influenced by seasonal conditions and crop choice.
For more information refer to Predicta B.
For more information see GRDC's Tips and tactics for crown rot.
For more information on rhizoctonia see GRDC's tips and tactics rhizoctonia.
For more information on root lesions nematodes refer to GRDC's Tips and Tactics Root Lesion nematodes.
Diagnosing Root disease
Predicta B
If a root disease needs to be identified there are two options. One method is the DNA based PreDicata B test which is conducted at SARDI (PIRSA). At present, the analyses provided by this service are only for soil pathogens using soil samples.
In most cases, a certified agronomist can collect the soil samples and obtain the results on behalf of growers for a fee.
Currently, the PreDicta B test can detect:
- Pratylenchus neglectus, P. quasiteriodes and P. thornei from the root lesion nematodes
- Stem nematode
- CCN
- Take-all (including the oat strain)
- Rhizoctonia solani
- Bipolaris (common root rot)
- Pythium clade
- Eyespot
- Crown rot
- Blackspot in field peas (PIRSA 2016).
Samples can be collected during the growing season, but soil samples are usually collected prior to sowing. This allows the grower to make informed decisions about disease management before planting crops. Results from the PreDicta B service include the disease risk levels of most of the detectable soil borne diseases as well as potential management options. PIRSA guarantee a one week turnaround for results.
DPIRD’s Diagnostic Laboratory Service (DDLS)
DDLS can provide analyses for root diseases as well as foliar diseases. The service can receive and process live plant matter as well as soil samples and separate charges apply for each disease requiring identification (Table 1). The most suitable analysis for the suspected disease(s) can be discussed with DDLS prior to submission of the samples. Unlike the PreDicta B service, the DDLS service does not offer information on potential disease risk or management and is more of a diagnostic tool.
For a full list of the services and associated fees click here.
Identifying and reporting exotic weeds, pests and diseases
Photos of potentially exotic weeds, pests and diseases can be sent directly to the Pest and Disease Identification Service (PaDIS) at DPIRD by email. If the photo does not give enough detail and it is suspected that the pest or disease is exotic to WA or a potential biosecurity threat, PaDIS may request a specimen be sent to the service. If the disease or pest is not exotic and the client requests identification from a specimen, charges will apply. See the website for details on what the PaDIS service includes and instructions for sample collection and delivery:
Pest and Disease Information Service (PaDIS) – DPIRD
Telephone: 08 9368 3080
Email: padis@dpird.wa.gov.au
Website: https://www.agric.wa.gov.au/biosecurity/pest-and-disease-information-service-padis




